
At this point we could decide that this is more than we are willing to try to patch, and leave the library bundled-not because of the build system, but because of the uthash 2.x incompatibility. * ucl_emitter_utils.c uses uthash internals, accessing the pd member directly, but this member no longer exists in 2.x * ucl_parser.c uses strtoimax without directly including inttypes.h, which was previously indirectly included from uthash.h this is also easily patched
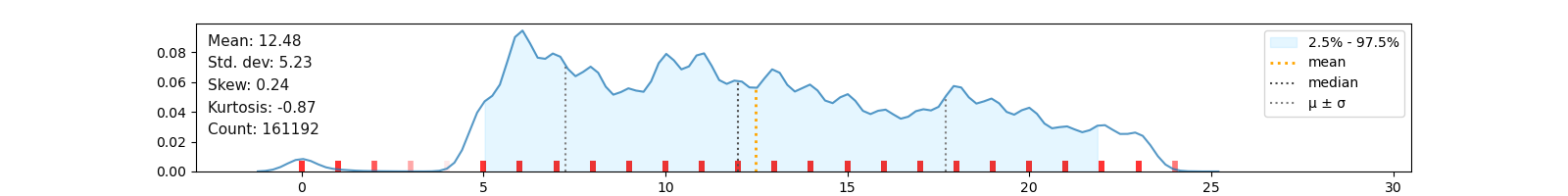
* libucl uses removed macros utstring_append_len() and utstring_append_c() this could be easily patched by defining them, if missing, in src/ucl_internal.h Now there are some problems with uthash 2.x, vs. Per the guidelines for depending on header-only libraries, you need to BR uthash-static in addition to uthash-devel. For example, for uthash, which was already in Fedora:ġ. It’s not hard to unbundle at least the header-only libraries. The linked SRPM is built with an old version of the spec file it does not match the spec URL. I kept the partially-bundled libraries in but marked themo so. The build system is a pain and does not seem to eat includes properly. The guidelines have not yet been updated. > An accepted change for Fedora 34 is that packages using make must BR it explicitly (“BuildRequires: make”). Regarding the above topic: I suppose I can remove `Cflags: -I$ check” would be better expressed as “%make_build check”. > Since /usr/include is a standard search path, explicitly adding it via -I can troublesome > in projects that want to customize the include directories. I don't think the 'packaging static library' guidelines are relevant here, since the package ships a dynamically linked library.? empty or not) checked for correlations among each other and with other features and in a second step for correlations with the label before a decision on ommitting them is made.I made the devel subpackage require arch-dependent base package, and run the test suite in the check section. Instead of simply dropping these columns, they are converted into binary features (i.e. Many parameters are available allowing a more restrictive data cleaning where needed.įurthermore, the function klib.mv_col_handling() provides a sophisticated selection mechanism for columns with relatively many missing values. Using this procedure, 56006 duplicate rows are identified in the subset, i.e., 56006 rows in 10 columns are encoded into a single column of dtype integer, greatly reducing the memory footprint and number of columns which should speed up model training.Īll of these functions were run with their relatively “soft” default settings. This allows us to pool and encode “carrier” and similar columns, while “tailnum” remains in the dataset.

While this is unlikely, it is advised to specifically exclude features that provide sufficient informational content by themselves as well as the target column by using the “exclude” setting.Īs can be seen in *cat_plot()* the “carrier” column is made up of a few very frequent values - the top 4 values account for roughly 75% - while in “tailnum” the top 4 values barely make up 2%. While the encoding itself does not lead to a loss in information, some details might get lost in the aggregation step. These are then added to the original data what allows dropping the previously identified and now encoded columns.
Specifically, the pooling is achieved by finding duplicates in subsets of the data and encoding the largest possible subset with sufficient duplicates with integers. This function “pools” columns together based on several settings. Further, klib.pool_duplicate_subsets() can be applied, what ultimately reduces the dataset to only 3.8 MB (from 51 MB originally).


 0 kommentar(er)
0 kommentar(er)
